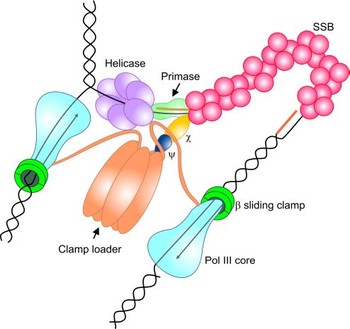
The information stored
in double-stranded DNA is sequestered in the interior of the protective double
helix. To provide access to genomic information, dsDNA must be unwound to form
single-stranded (ss) intermediates. Since ssDNA is prone to chemical and
nucleolytic attacks this process is risky since it can cause damage that is
difficult to repair. To help preserve ssDNA intermediates, cells have evolved a
specialized class of ssDNA-binding (SSB) proteins that associate with ssDNA
with high affinity and in a sequence-independent manner. Beyond their roles in
DNA binding, SSB proteins do also associate with a broad array of cellular
genome maintenance proteins. This interaction in many cases stimulates the
biochemical activities of SSB's partner proteins.
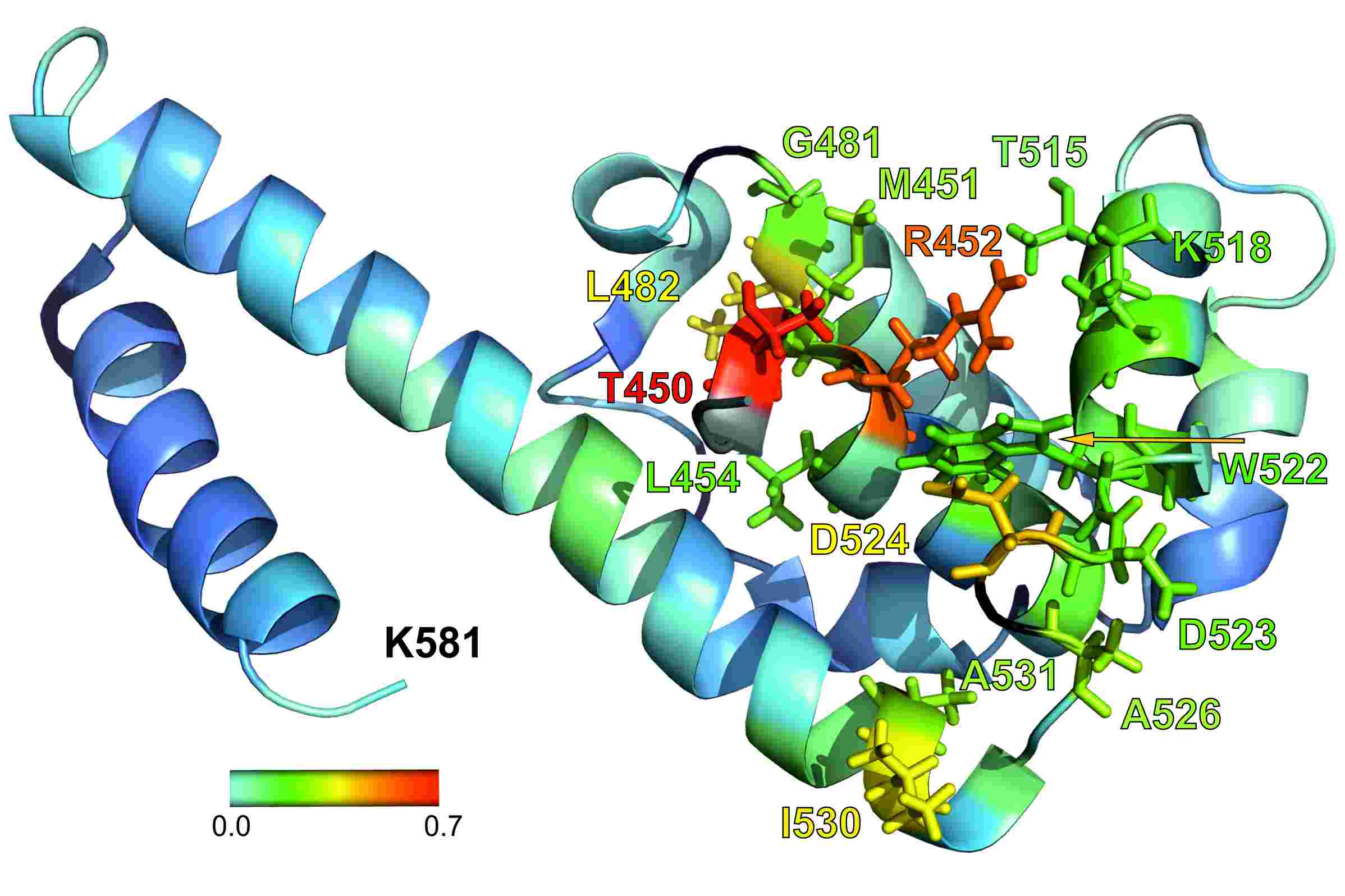
We are using NMR-spectroscopy in conjunction with several
labeling pattern to elucidate the interaction of SSB proteins, in particular
the C-terminus of SSB proteins, with various interactions partners in the the
bacterial replication fork.